We have a new paper out in eLife: Uncovering the electrical synapse proteome in retinal neurons via in vivo proximity labeling.
Authors: Stephan Tetenborg, Eyad Shihabeddin, Elizebeth Olive Akansha Manoj Kumar, Crystal Sigulinsky @csigulinsky.bsky.social, Karin Dedek, Ya-Ping Lin, Fabio Echeverry, Hannah Hoff, Alberto Pereda, Bryan William Jones @bwjones.bsky.social, Christophe Ribelayga, Klaus Ebnet, Ken Matsuura, John O’Brien
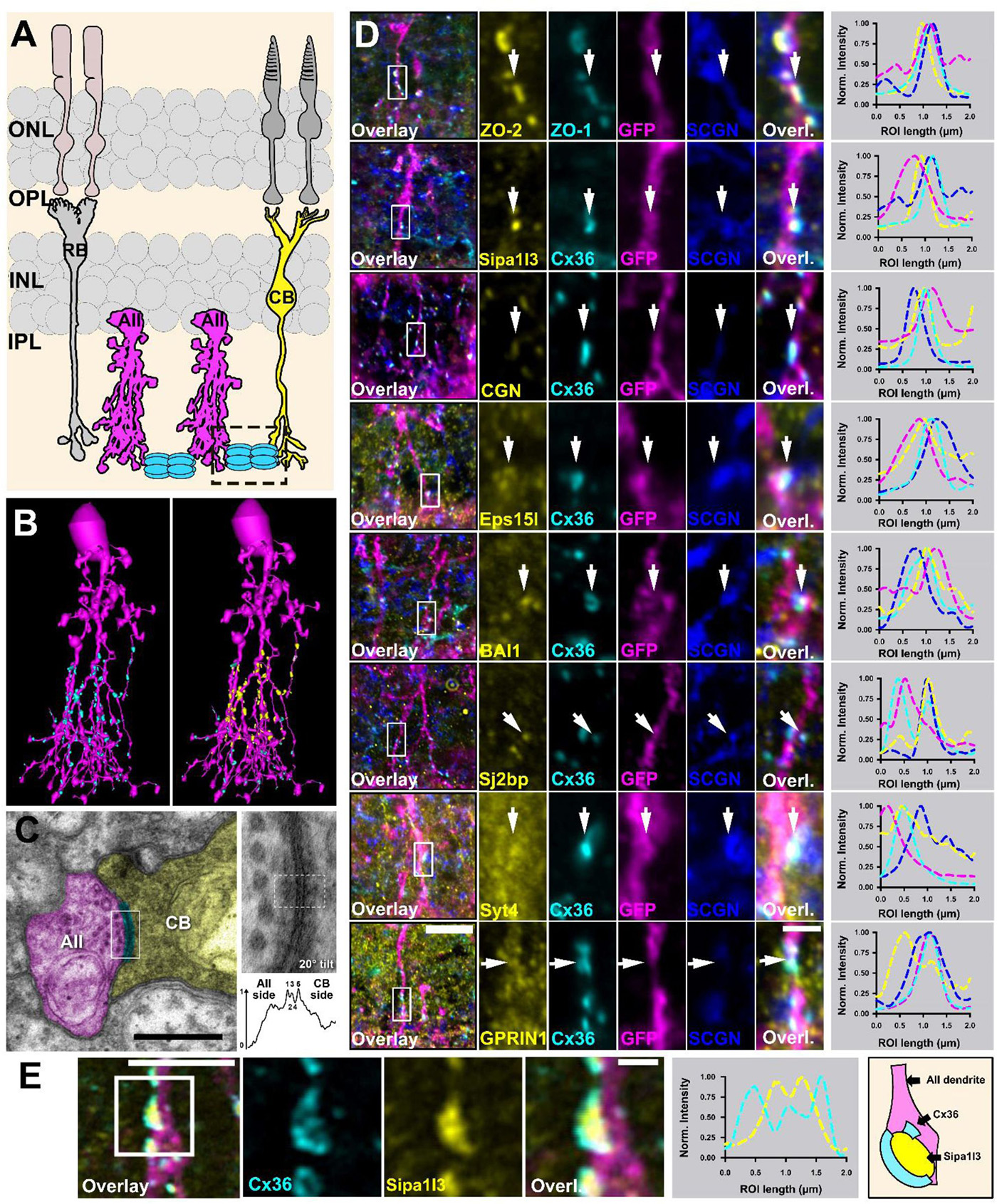
Abstract: Electrical synapses containing Connexin 36 (Cx36) represent the main means for direct electrical communication among neurons in the mammalian nervous system. However, little is known about the protein complexes that constitute these synapses. In the present study, we applied different BioID strategies to screen the interactomes of Connexin 36 and its zebrafish orthologue Cx35b in retinal neurons. For in vivo proximity labeling in mice, we took advantage of the Cx36-EGFP strain and expressed a GFP-nanobody-TurboID fusion construct selectively in AII amacrine cells. For in vivo BioID in zebrafish, we generated a transgenic line expressing a Cx35b-TurboID fusion under control of the Cx35b promoter. Both strategies allowed us to capture a plethora of molecules that were associated with electrical synapses and showed a high degree of evolutionary conservation in the proteomes of both species. Besides known interactors of Cx36 such as ZO-1 and ZO-2 we have identified more than 50 new proteins, such as scaffold proteins, adhesion molecules and regulators of the cytoskeleton. Moreover, we determined the subcellular localization of these proteins in mouse retina and tested potential binding interactions with Cx36. Amongst these new interactors, we identified signal induced proliferation associated 1 like 3 (Sipa1l3), a protein that has been implicated in cell junction formation and cell polarity, as a new scaffold of electrical synapses. Interestingly, Sipa1l3 was able to interact with ZO-1, ZO-2 and Cx36, suggesting a pivotal role in electrical synapse function. In summary, our study provides the first detailed view of the electrical synapse proteome in retinal neurons, which is likely to apply to electrical synapses elsewhere.